Designer Enzymes
Ongoing projects in the Hilvert lab focus on design and directed evolution of enzymes with tailormade properties. Our work aims at expanding the scope of protein biocatalysis towards non-natural chemical transformations, either by modifying natural enzymes or by rational design of artificial ones.
Over the last two decades, advances in computational methodologies have made it possible to generate de novo enzymes that catalyze a broad range of abiological reactions. These design efforts are typically combined with experimental optimization of the respective catalysts by directed evolution, where iterative cycles of mutagenesis and screening or selection are used to achieve substantial improvements in catalytic efficiency, stability and stereoselectivity. Such systems are valuable as tools for mechanistic enzymology, and may also be customized for diverse biocatalytic applications.
We apply medium- and high-throughput screening techniques to optimize various classes of computationally designed enzymes, including esterases, Diels-Alderases, Kemp eliminases, and aldolases. Detailed structural, kinetic and biochemical characterization of the evolved catalysts provides insight into underlying enzyme mechanisms as well as crucial feedback for the computational design process.
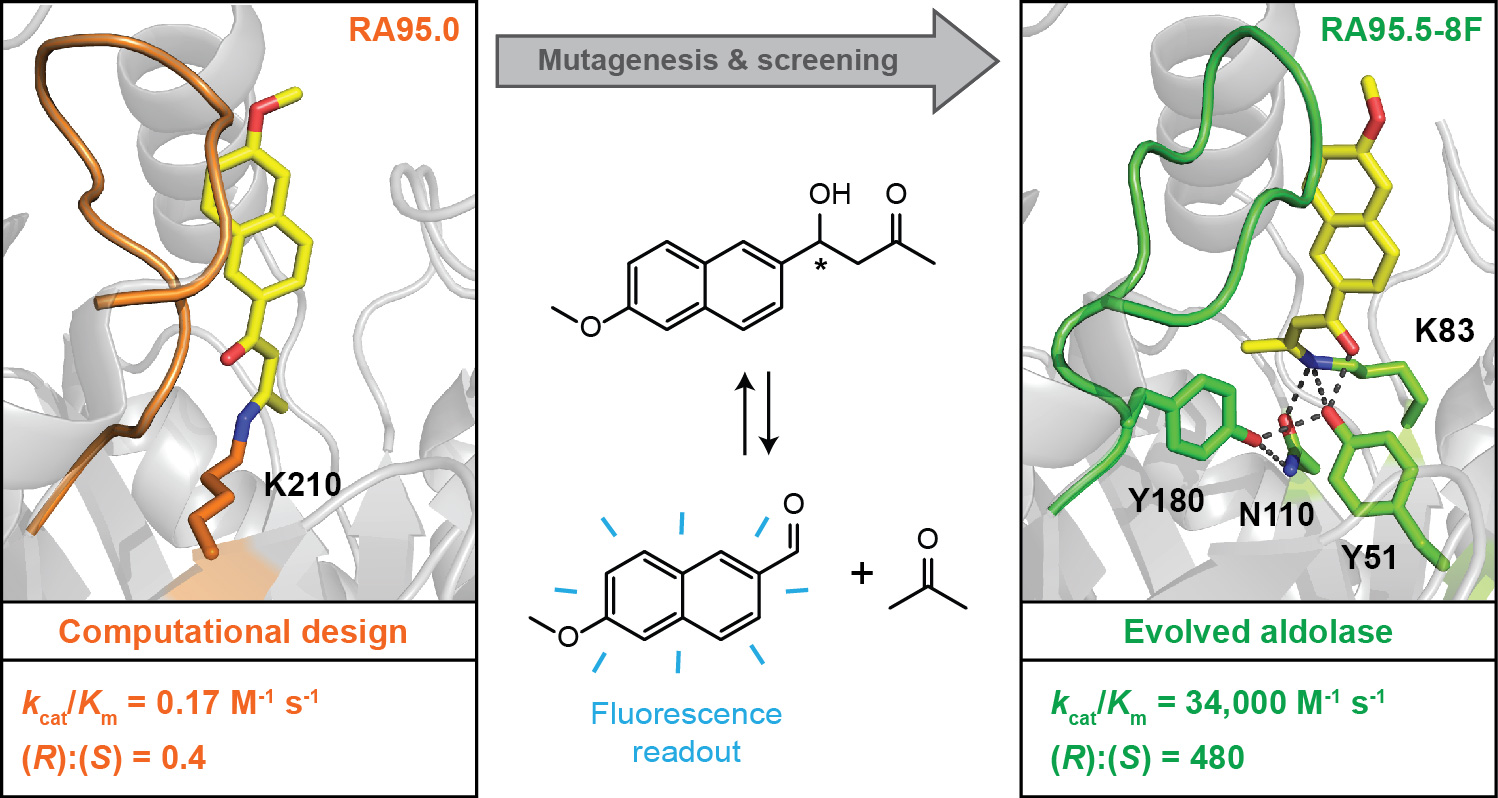
In one recent example, we improved the catalytic efficiency of a de novo aldolase (RA95) by five orders of magnitude using conventional microtiter plate screening for the initial rounds and ultrahigh-throughput droplet-based microfluidic screening during the late stages of directed evolution (Figure 1). Over the course of optimization, a sophisticated hydrogen-bond network emerged at the active site that facilitates Schiff base formation and mechanistically important proton transfers along the multistep reaction coordinate. Furthermore, the promiscuity of the enzyme has been exploited for mechanistically related reactions, such as Michael additions, Henry reactions and Knoevenagel condensations.
These efforts have recently been extended to de novo enzymes based on naïve artificial protein scaffolds, which provide customized and unbiased starting points for the design and evolution of novel biocatalysts. We are also designing and evolving artificial metalloenzymes, where the versatile reactivity of a metal cofactor is combined with the superb specificity of a chiral active site in a protein.
In an alternative approach, we are applying genetic code expansion to tailor the properties of natural heme proteins, such as ascorbate peroxidase and myoglobin. Mutation of the proximal heme ligand to the non-canonical amino acid N-methyl histidine, for example, fine-tunes the heme iron’s reduction potential and serves as a valuable mechanistic probe. Directed evolution can then be employed to optimize these engineered heme proteins for catalysis of various reactions. For instance, myoglobin’s promiscuous peroxidase activity increased substantially by introducing N-methyl histidine and further optimization by several rounds of laboratory evolution led to variants with catalytic efficiencies comparable to those of natural peroxidases. Such catalysts are also useful for accelerating abiological carbene transfer reactions.
Selected Publications
Studer, S., Hansen, D. A., Pianowski, Z. L., Mittl, P. R., Debon, A., Guffy, S. L., Der, B. S., Kuhlman, B., Hilvert, D. Evolution of a highly active and enantiospecific metalloenzyme from short peptides. Science, 362, 1285 (2018).
Hayashi, T., Tinzl, M., Mori, T., Krengel, U., Proppe, J., Soetbeer, J., Klose, D., Jeschke, G., Reiher, M., Hilvert, D. Capture and characterization of a reactive heme-carbenoid in an artificial metalloenzyme. Nat. Catal., 1, 578 (2018).
Pott, M., Hayashi, T., Mori, T., Mittl, P. R., Green, A. P., Hilvert, D. A noncanonical proximal heme ligand affords an efficient peroxidase in a globin fold. J. Am. Chem. Soc., 140, 1535 (2018).
Garrabou, X., Macdonald, D. S., Wicky, B. I., Hilvert, D. Stereodivergent evolution of artificial enzymes for the Michael reaction. Angew. Chem. Int. Ed., 57, 5288 (2018).
Zeymer, C., Zschoche, R., Hilvert, D. Optimization of enzyme mechanism along the evolutionary trajectory of a computationally designed (retro-)aldolase. J. Am. Chem. Soc., 139, 12541 (2017).
Obexer, R., Godina, A., Garrabou, X., Mittl, P. R., Baker, D., Griffiths, A. D., Hilvert, D. Emergence of a catalytic tetrad during evolution of a highly active artificial aldolase. Nat. Chem., 9, 50 (2017).
Garrabou, X., Macdonald, D. S., Hilvert, D. Chemoselective Henry condensations catalysed by artificial carboligases. Chem. Eur. J., 23, 6001 (2017).
Obexer, R., Pott, M., Zeymer, C., Griffiths, A. D., Hilvert, D. Efficient laboratory evolution of computationally designed enzymes with low starting activities using fluorescence-activated droplet sorting. Protein. Eng. Des. Sel., 29, 355 (2016).
Green, A. P., Hayashi, T., Mittl, P. R., Hilvert, D. A chemically programmed proximal ligand enhances the catalytic properties of a heme enzyme. J. Am. Chem. Soc., 138, 11344 (2016).
Garrabou, X., Wicky, B. I., Hilvert, D. Fast Knoevenagel condensations catalyzed by an artificial Schiff-base-forming enzyme. J. Am. Chem. Soc., 138, 6972 (2016).
Garrabou, X., Beck, T., Hilvert, D. A promiscuous de novo retro-aldolase catalyzes asymmetric Michael additions via Schiff base intermediates. Angew. Chem. Int. Ed., 54, 5609 (2015).
Preiswerk, N., Beck, T., Schulz, J. D., Milovnik, P., Mayer, C., Siegel, J. B., Baker, D., Hilvert, D. Impact of scaffold rigidity on the design and evolution of an artificial Diels-Alderase. Proc. Natl. Acad. Sci. U. S. A., 111, 8013 (2014).
Blomberg, R., Kries, H., Pinkas, D. M., Mittl, P. R., Grutter, M. G., Privett, H. K., Mayo, S. L., Hilvert, D. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature, 503, 418 (2013).
Giger, L., Caner, S., Obexer, R., Kast, P., Baker, D., Ban, N., Hilvert, D. Evolution of a designed retro-aldolase leads to complete active site remodeling. Nat. Chem. Biol., 9, 494 (2013).